SCIAMA
High Performance Compute Cluster
Using Spyder on SCIAMA
Spyder is installed on SCIAMA as a package of cpython/3.8.9, load module cython/3.8.9. You will need to be using a GUI: x2Go has been installed on nodes login3, login4, login5 and login6.
Using Kernels launched on compute nodes:
From the command line on a login node run sinteractive; this will start an interactive session on a compute node.
Once on the compute node, to start a kernel type:
You should then see the kernel name as shown here:
To exit, you will have to explicitly quit this process, by either sending "quit" from a client, or using Ctrl-\ in UNIX-like environments.
To read more about this, see https://github.com/ipython/ipython/issues/2049
To connect another client to this kernel, use:
--existing kernel-51072.json
In Spyder you will need to connect to the kernel:
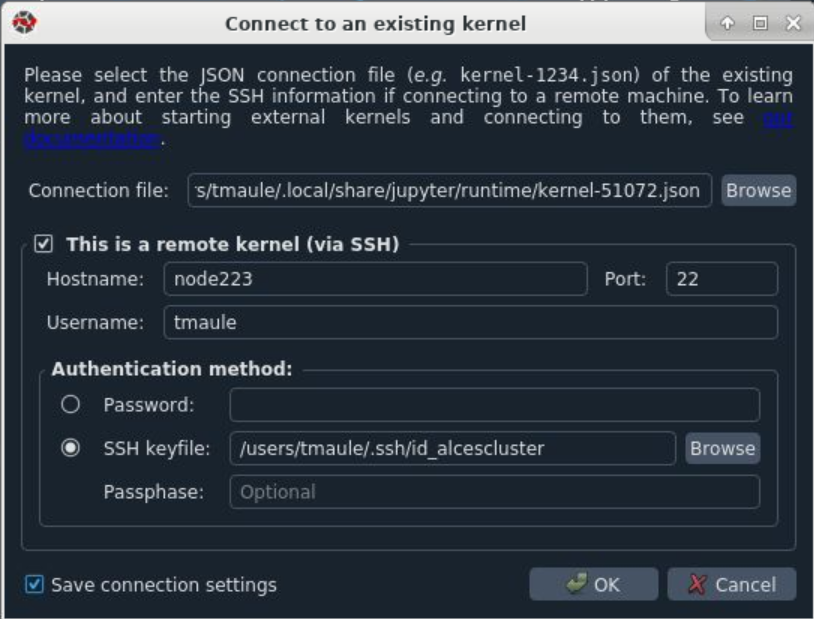
Go to Consoles --> Connect to an existing kernel (this may take a few minutes to launch)
In the Connection file: field browse to the kernel you have started on the compute node. In my case it is kernel-570172.json
Tick the box for "This is a remote kernel (via ssh)" and add the Hostname of the same node you launched your kernel, again in my case this is node223. Enter your username.
Under "Authentication Method:" use SSH keyfile: and type in .ssh/id_alcescluster. No passphrase is needed so leave it blank. Tick the Save connection settings box and hit ok.
This will launch a new console and connect to the kernel. When you run your python script it should run in the console on the compute node.
In theory you can launch as many kernels as you like on different compute nodes.